
Estructura de datos de imágenes médicas ampliada a múltiples modalidades y regiones anatómicas
J. M. Saborit-Torres, J. À. Montell, J. M. Salinas, J. A. Gómez, I. Stefan, M. Caparrós, F. García-García, J. Domenech, J. V. Manjón, G. Rojas, A. Pertusa, A. Bustos, G. González, J. Galant, M. de la Iglesia-Vayá
Resumen
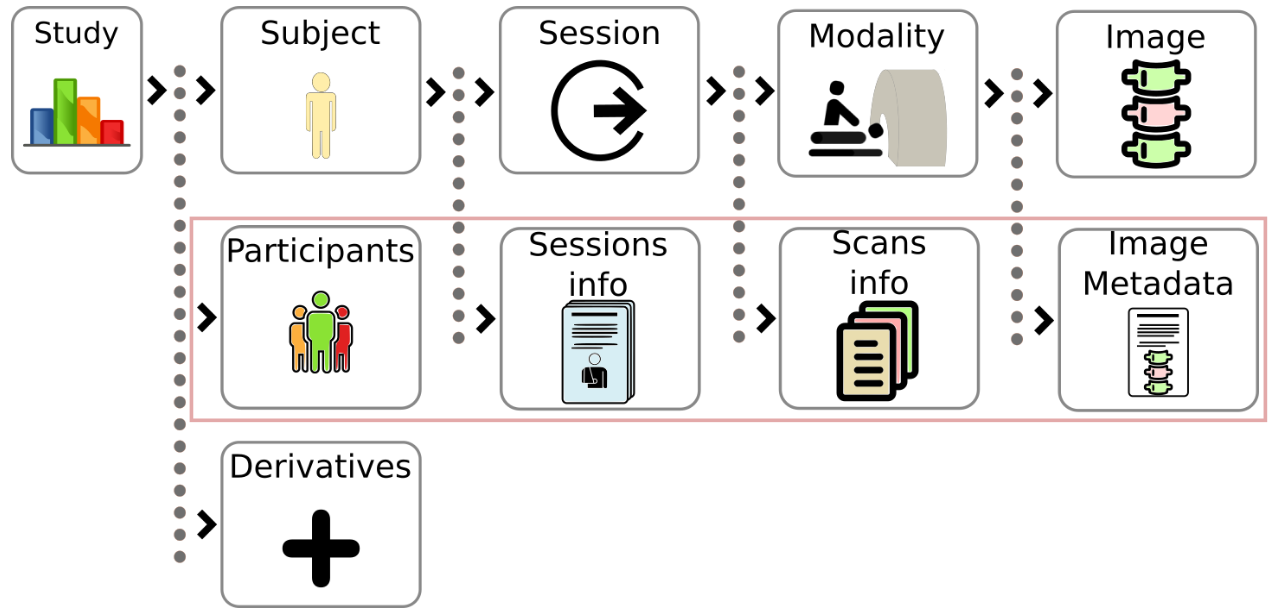
Brain Imaging Data Structure (BIDS) permite al usuario organizar los datos de imágenes cerebrales en una estructura de directorios estándar clara y sencilla. BIDS cuenta con un amplio respaldo de la comunidad científica y se considera un potente estándar de gestión. El BIDS original se limita a imágenes o datos relacionados con el cerebro. Por ello, Medical Imaging Data Structure (MIDS) se concibió con el objetivo de ampliar esta metodología a otras regiones anatómicas y a otros tipos de sistemas de obtención de imágenes en estas áreas.
Palabras clave: normalización – ampliación – estructura de datos de imágenes cerebrales – estructura de datos de imágenes médicas – base de datos – región anatómica – modalidad
Software
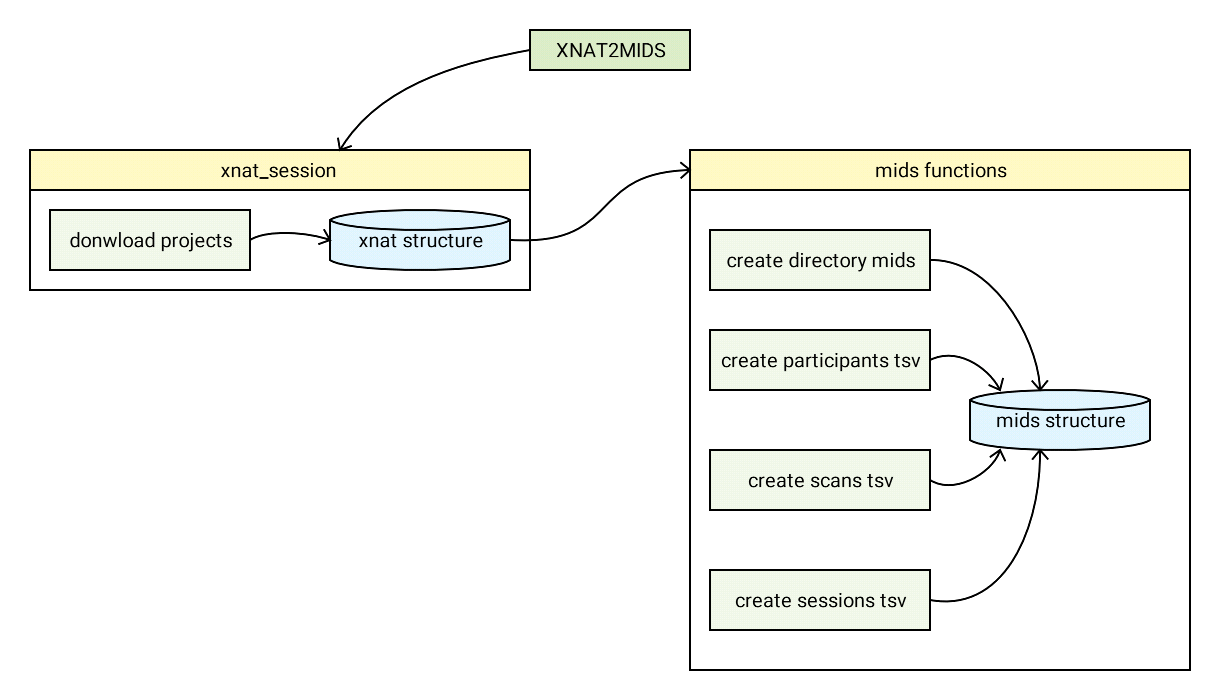
XNAT2MIDS es un software escrito en Python 3 y puede encontrarse en el repositorio público de Github arriba indicado. Este software permite a los usuarios de una plataforma XNAT conectarse a sus proyectos asignados y descargar los proyectos de interés en formato MIDS. La siguiente figura muestra el flujo de ejecución en el que se genera una sesión XNAT. En ella se pueden almacenar todos los proyectos solicitados y, una vez guardados en un directorio temporal, el programa genera la estructura general del MIDS y sus metadatos tabulares.
Datos
Esta estructura de directorios se aplicó a los datos del banco de imágenes médicas de la Comunidad Valenciana en el proyecto BIMCV COVID19 +. Este proyecto es un gran conjunto de datos del Banco de Datos de Imágenes Médicas en BIMCV con imágenes de rayos X de tórax CXR (CR, DX) y tomografía computarizada (TC) de imágenes de SARS-CoV-2 (COVID 19).
Contacto
Si quiere saber más sobre el proyecto o ponerse en contacto con el equipo de investigación, escríbanos.

